This tutorial is for NSD-iEEG dataset (link is to be added when the paper is published). For more information on the Natural Scenes Dataset (NSD), please check out website (https://naturalscenesdataset.org).
Data and program setups
NSD-iEEG in BIDS Format
First, please download the NSD-iEEG data prepared in BIDS format. Here is an example file layout and critical folders. Make sure the project name is called NSDiEEG.
NSDiEEG/ <- root directory
├─sub-01
│ └─ses-ieeg01/
│ └─ieeg/
| └─sub-01_ses-ieeg01_electrodes.tsv <- electrode coordinates in T1w
├─sub-02/
└─derivatives/
└─freesurfer/
├─sub-01/ <- Subject's FreeSurfer directory
│ ├─mri/
│ ├─surf/
│ ├─label
│ └─...
└─sub-02/Make sure RAVE is up-to-date
Please make sure RAVE is installed. If you have installed prior to May 22, 2025 and haven’t updated since then, you need to update RAVE
The RAVE pipeline to visualize the NSD-iEEG data is published on Github. You can use RAVE’s built-in functions to load the script:
# Make sure the pipeline is up-to-date
ravepipeline::update_local_snippet()Generate subject-level 3D viewer
To generate a 3D viewer for a NSD-iEEG subject, only two parameters are needed
bids_project_path: absolute path to the BIDS dataset root directory. You can verify the path by checking whether filedatasets_description.jsonexists under the pathbids_subject_code: subject code, with or without the leadingsub-
For example, here is how to generate a standalone 3D viewer for sub-06:
import_nsd_ieeg = ravepipeline::load_snippet("import-nsd-electrodes")
result = import_nsd_ieeg(
bids_project_path = "~/rave_data/nsd_data/NSDiEEG/",
bids_subject_code = "06")
#> Migrating `freesurfer` directory with overwrite: FALSE
#> Source: /Users/zj/rave_data/nsd_data/NSDiEEG/derivatives/freesurfer/sub-06
#> Destination: /Users/zj/rave_data/raw_dir/NSDiEEG_06/rave-imaging/fs
#> Found the following electrode coordinate files:
#> parsed data_type suffix extension sub ses space
#> <AsIs> <char> <char> <char> <char> <char> <char>
#> 1: sub-06/s.... ieeg electrodes tsv 06 ieeg01 <NA>
#> 2: sub-06/s.... ieeg electrodes tsv 06 ieeg01 MNI152NLin2009
#> 3: sub-06/s.... ieeg electrodes tsv 06 ieeg01 MNI305
#> Using BIDS tabular to generate RAVE electrodes.csv:
#> /Users/zj/rave_data/nsd_data/NSDiEEG/./sub-06/ses-ieeg01/ieeg/sub-06_ses-ieeg01_electrodes.tsv
#> Compute T1w scanner RAS -> FreeSurfer surface coordinates...
#> `Destrieux_label_text` found... using existing labeling.
#> Found MNI152 from BIDS tabular
#> /Users/zj/rave_data/nsd_data/NSDiEEG/./sub-06/ses-ieeg01/ieeg/sub-06_ses-ieeg01_space-MNI152NLin2009_electrodes.tsv
#> Saving the electrode coordinates
#> Computing mapping to fsaverage by projecting contacts to surface: smoothwm
#> Saving to `rave/meta/electrodes.csv` with MNI152 and fsaverage mapping.
#> Saving the viewer to:
#> /Users/zj/rave_data/nsd_data/NSDiEEG/derivatives/rave/sub-06/exports/viewers/nsd_import.html
print(result)
#> $subject
#> [1] "NSDiEEG/NSDiEEG_06"
#>
#> $electrode_path
#> [1] "/Users/zj/rave_data/data_dir/NSDiEEG/NSDiEEG_06/rave/meta/electrodes.csv"
#>
#> $viewer_path
#> [1] "/Users/zj/rave_data/nsd_data/NSDiEEG/derivatives/rave/sub-06/exports/viewers/nsd_import.html"RAVE subject NSDiEEG_06 will be created with the following data:
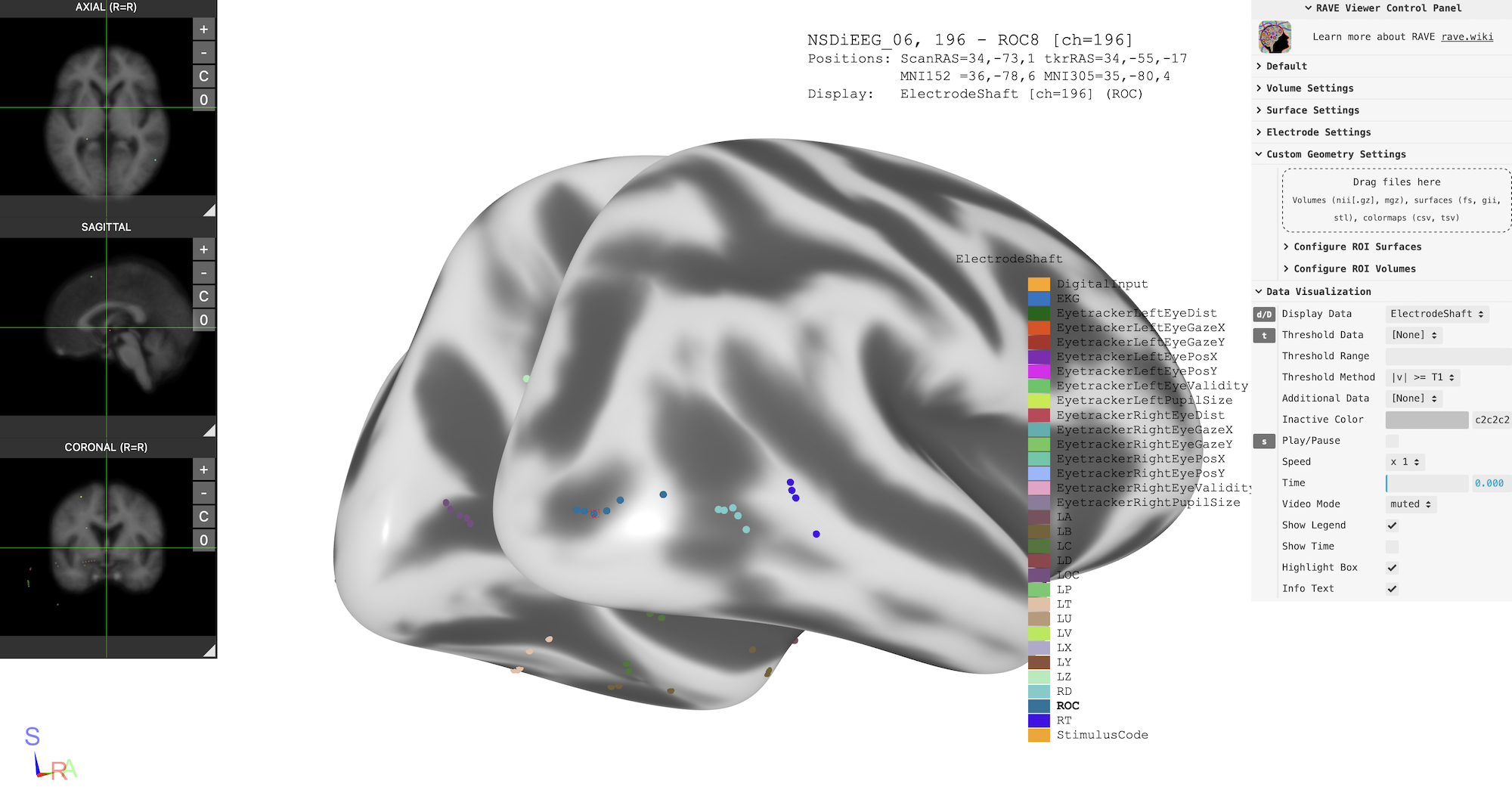
electrodes.csv: this is the electrode coordinates used by RAVE, that contains electrode coordinates in T1 scanner, FreeSurfer tk-registered RAS, MNI-152, and FreeSurfer spherical coordinatesnsd_import.html: Individual 3D viewer; this viewer is standalone and can be shared and opened with no dependency (you just need a web-browser with 3D graphics support)
Here is an example of the subject viewer with a few “tweak” (from control panel, turn on Electrode Settings -> Map Electrodes", then choose "inflated" brain surface fromSurface Settings -> Surface Type`)
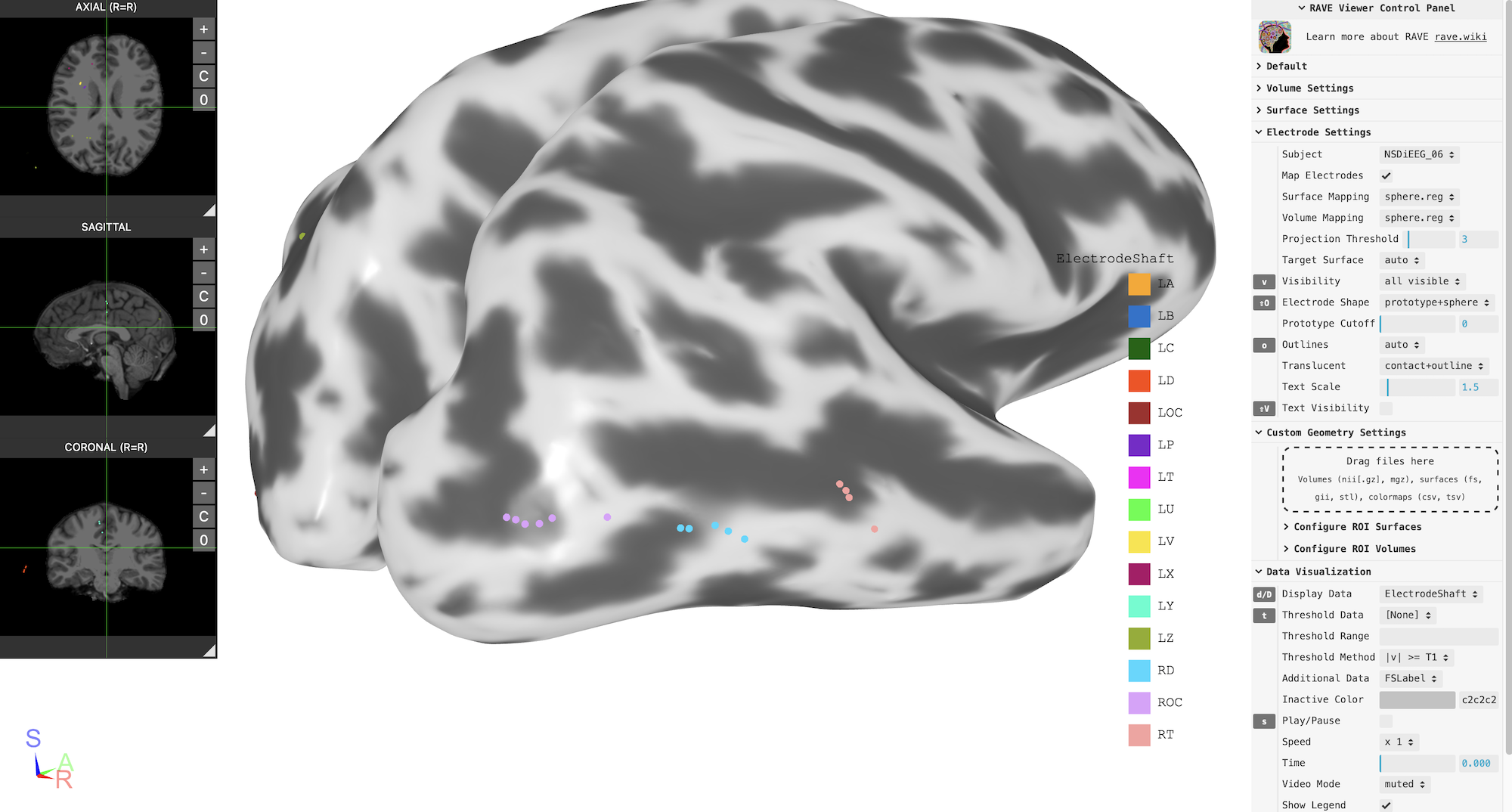
Group-level viewer
Once the subject is imported to RAVE, you can use RAVE to generate group-level viewer on many popular templates. Here is an example using fsaverage. Other choices are cvs_avg35_inMNI152 (MNI152), N27 (Collin’s brain)
To start, let’s make sure you have the template. This is one-time setup code.
threeBrain::download_template_subject("fsaverage")Next, load the brain for all the subjects from NSD-iEEG data
# ---- inputs --------------
# You can add more subjects here
subjects = c("06")
template = "fsaverage"
# ---- code ----------------
# load each subject brain
brain_list = list()
for(subject_code in subjects) {
rave_subject = raveio::as_rave_subject(sprintf("NSDiEEG/NSDiEEG_%s", subject_code))
brain = raveio::rave_brain(rave_subject, surfaces = c("sphere.reg"))
brain$set_electrode_values()
brain_list[[length(brain_list) + 1]] = brain
}
# load template brain
template_brain = threeBrain::merge_brain(.list = brain_list, template_subject = template, template_surface_types = c("inflated", "sphere.reg"))
template_brain$plot(
controllers = list(
"Display Data" = "ElectrodeShaft",
"Volume Mapping" = "sphere.reg",
"Projection Threshold" = 3
)
)